Data radtorch.data
One of the core functions of radtorch is the ability to handle different types of medical/non-medical DICOM/non-DICOM images efficiently with ease. Below is list of classes that make the magic happen.
ImageObject
Creates a 3D tensor whose dimensions = [channels, width, height] from an image path.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
path |
str |
Path to an image. |
required |
out_channels |
int |
Number of output channels. Only 1 and 3 channels supported. |
required |
transforms |
list |
Albumentations transformations. See Image Augmentation. |
required |
WW |
int or list |
Window width for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
required |
WL |
int or list |
Window level for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
required |
Returns:
| Type | Description |
|---|---|
tensor |
3D tensor whose dimensions = [channels, width, height] |
Examples:
>>> i = radtorch.data.ImageObject(path='data/PROTOTYPE/DIRECTORY/abdomen/abd_1/1-001.dcm')
>>> i.shape
torch.Size([1, 512, 512])
VolumeObject
Creates an Image Volume Object (4D tensor) from series images contained in a folder.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
directory |
str |
Folder containing series/sequence images. Images must be DICOM files. |
required |
out_channels |
int |
Number of output channels. Only 1 and 3 channels supported. |
required |
transforms |
list |
Albumentations transformations. See https://albumentations.ai/docs/getting_started/image_augmentation/. |
required |
WW |
int or list |
Window width for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
required |
WL |
int or list |
Window level for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
required |
Returns:
| Type | Description |
|---|---|
tensor |
4D tensor with dimensions = [channels, number_images/depth, width, height]. See https://pytorch.org/docs/stable/generated/torch.nn.Conv3d.html |
Examples:
>>> i = radtorch.data.VolumeObject(directory='data/PROTOTYPE/DIRECTORY/abdomen/abd_1')
>>> i.shape
torch.Size([1, 40, 512, 512])
ImageDataset (Dataset)
Creates pytorch dataset(s) and dataloader(s) objects from a parent folder. Use this class for image tasks that invovles handling each single image as a single instance of your dataset.
Examples:
import radtorch
import albumentations as A
# Specify image transformations
T = A.Compose([A.Resize(256,256)])
# Create dataset object
ds = radtorch.data.ImageDataset(
folder='data/4CLASS/',
split={'valid':0.2, 'test':0.2},
out_channels=1,
transforms={'train':T,'valid': T,'test': T},
)
ds.data_stat()
ds.table

Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
folder |
str |
Parent folder containing images. |
required |
name |
str |
Name to be give to the dataset. If none provided, the current date and time will be used to created a generic dataset name. (default=None) |
None |
label_table |
str|dataframe |
The table containing data labels for your images. Expected table should contain at least 2 columns: image path column and a label column. Table can be string path to CSV or a pandas dataframe object.(default=None) |
None |
instance_id |
bool |
True if the data provided in the image path column in label_table contains the image id not the absolute path for the image. (default= False) |
False |
add_extension |
bool |
If instance_id =True, use this to add extension to image path as needed. Extension must be provided without "." e.g. "dcm". (default=False) |
False |
out_channels |
int |
Number of output channels. (default=1) |
1 |
WW |
int or list |
Window width for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
None |
WL |
int or list |
Window level for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
None |
path_col |
str |
Name of the column containing image path data in the label_table. (default='path') |
'path' |
label_col |
str |
Name of the column containing label data in the label_table. (default='label') |
'label' |
extension |
str |
Type/Extension of images. (default='dcm') |
'dcm' |
transforms |
dict |
Dictionary of Albumentations transformations in the form of {'train': .. , 'valid': .. , 'test': .. }. See https://albumentations.ai/docs/getting_started/image_augmentation/ . (default=None) |
None |
random_state |
int |
Random seed (default=100) |
100 |
sample |
float |
Sample or percent of the overall data to be used. (default=1.0) |
1.0 |
split |
dict |
dictionary defining how data will be split for training/validation/testing. Follows the sturcture {'valid': float, 'test': float} or {'valid':'float'} in case no testing subset is needed. The percent of the training subset is infered automatically. |
False |
ignore_zero_img |
bool |
True to ignore images containig all zero pixels. (default=False) |
False |
normalize |
bool |
True to normalize image data between 0 and 1. (default=True) |
True |
batch_size |
int |
Dataloader batch size. (default = 16) |
16 |
shuffle |
bool |
True to shuffle images during training. (default=True) |
True |
weighted_sampler |
bool |
True to use a weighted sampler for unbalanced datasets. See https://pytorch.org/docs/stable/data.html#torch.utils.data.WeightedRandomSampler. (default=False) |
False |
num_workers |
int |
Dataloader CPU workers. (default = 0) |
0 |
Attributes:
| Name | Type | Description |
|---|---|---|
classes |
list |
List of generated classes/labels. |
class_to_idx |
dict |
Dictionary of generated classes/labels and corresponding class/label id. |
idx_train |
list |
List of index values of images/instances used for training subset. These refer to index of |
idx_valid |
list |
List of index values of images/instances used for validation subset. These refer to index of |
idx_test |
list |
List of index values of images/instances used for testing subset. These refer to index of |
table |
pandas dataframe |
Table of images , paths and their labels. |
table_train |
pandas dataframe |
Table of images used for training. Subset of |
table_valid |
pandas dataframe |
Table of images used for validation. Subset of |
table_test |
pandas dataframe |
Table of images used for testing. Subset of |
tables |
dict |
Dictionary of all generated tables in the form of: {'train': table, 'valid':table, 'test':table}. |
dataset_train |
pytorch dataset object |
Training pytorch Dataset |
dataset_valid |
pytorch dataset object |
Validation pytorch Dataset |
dataset_test |
pytorch dataset object |
Testing pytorch Dataset |
datasets |
dict |
Dictionary of all generated Datasets in the form of: {'train': Dataset, 'valid':Dataset, 'test':Dataset}. |
dataloader_train |
pytorch dataloader object |
Training pytorch DataLoader |
dataloader_valid |
pytorch dataloader object |
Validation pytorch DataLoader |
dataloader_test |
pytorch dataloader object |
Testing pytorch DataLoader |
dataloaders |
dict |
Dictionary of all generated Dataloaders in the form of: {'train': Dataloader, 'valid':Dataloader, 'test':Dataloader}. |
class_weights |
tensor |
Values of class weights, for imbalanced datasets, to be used to weight loss functions. See https://pytorch.org/docs/stable/generated/torch.nn.CrossEntropyLoss.html#torch.nn.CrossEntropyLoss. |
sampler_weights |
tensor |
Values of vlass weights, for imbalanced datasets, to be used to sample from the dataset using Pytroch WeightedRandomSampler. Affects only training dataset not validation or testing. See https://pytorch.org/docs/stable/data.html#torch.utils.data.WeightedRandomSampler |
Methods
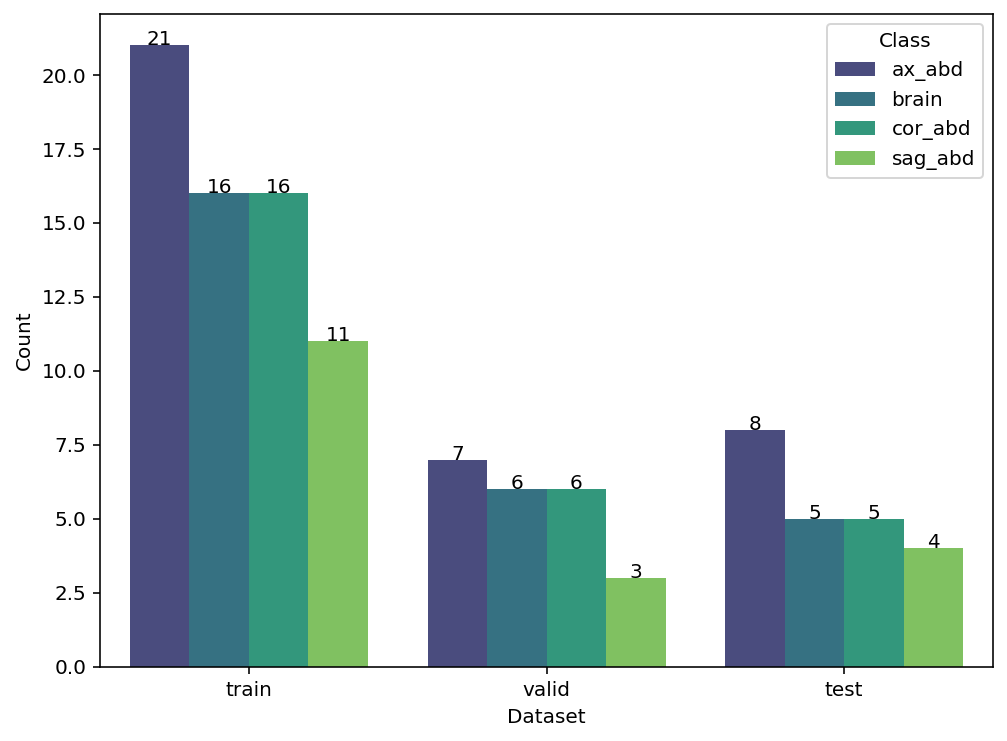
data_stat(self, plot=True, figsize=(8, 6), cmap='viridis')
Displays distribution of classes across subsets as table or plot.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
plot |
bool |
True, display data as figure. False, display data as table. |
True |
figsize |
tuple |
size of the displayed figure. |
(8, 6) |
cmap |
string |
Name of Matplotlib color map to be used. See Matplotlib colormaps |
'viridis' |
Returns:
| Type | Description |
|---|---|
pandas dataframe |
if plot=False |
Examples:
ds = radtorch.data.ImageDataset(folder='data/4CLASS/', out_channels=1)
ds.data_stat()
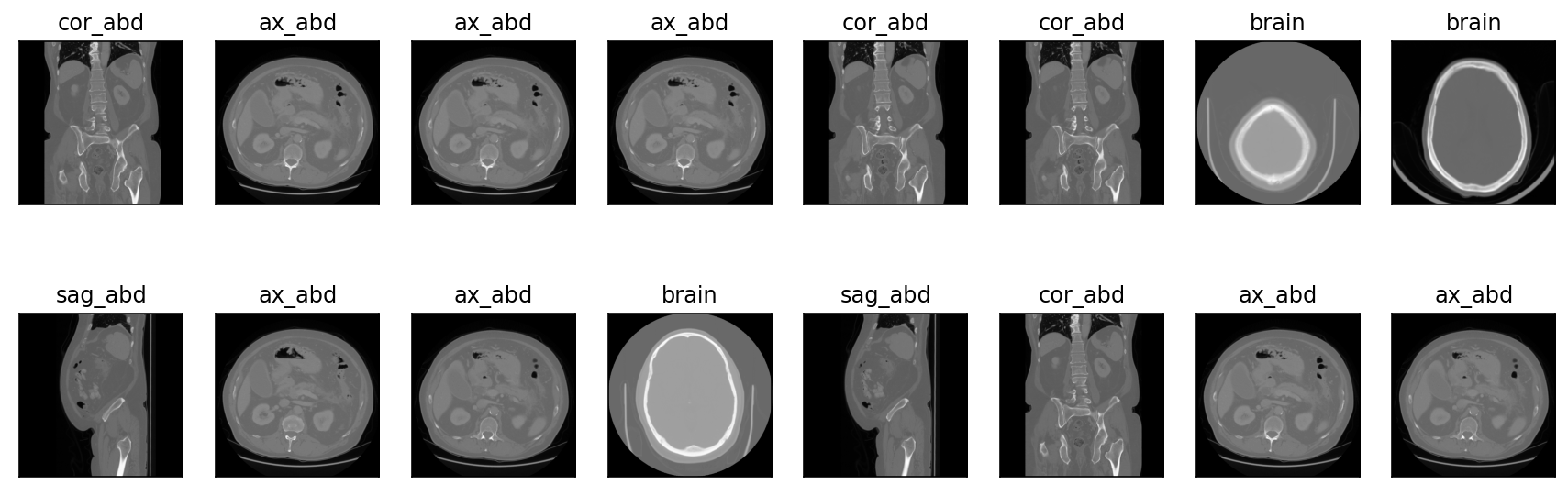
view_batch(self, subset='train', figsize=(15, 5), cmap='gray', num_images=False, rows=2)
Displays a batch from a certain subset.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
subset |
string |
Datasubset to use: either 'train', 'valid', or 'test'. |
'train' |
figsize |
tuple |
Size of the displayed figure. |
(15, 5) |
cmap |
string |
Name of Matplotlib color map to be used. See Matplotlib colormaps |
'gray' |
num_images |
int |
Number of displayed image. Usually equals batch_size unless otherwise specified. |
False |
rows |
int |
Number of rows. |
2 |
Returns:
| Type | Description |
|---|---|
figure |
figure containing samples |
Examples:
ds = radtorch.data.ImageDataset(folder='data/4CLASS/', out_channels=1)
ds.view_batch()
view_image(self, id=0, figsize=(25, 5), cmap='gray')
Displays separate images/channels of an image.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
id |
int |
Target image id in |
0 |
figsize |
tuple |
size of the displayed figure. |
(25, 5) |
cmap |
string |
Name of Matplotlib color map to be used. See Matplotlib colormaps |
'gray' |
Returns:
| Type | Description |
|---|---|
figure |
figure containing samples |
Examples:
ds = radtorch.data.ImageDataset(folder='data/4CLASS/', out_channels=3)
ds.view_image(id=9)
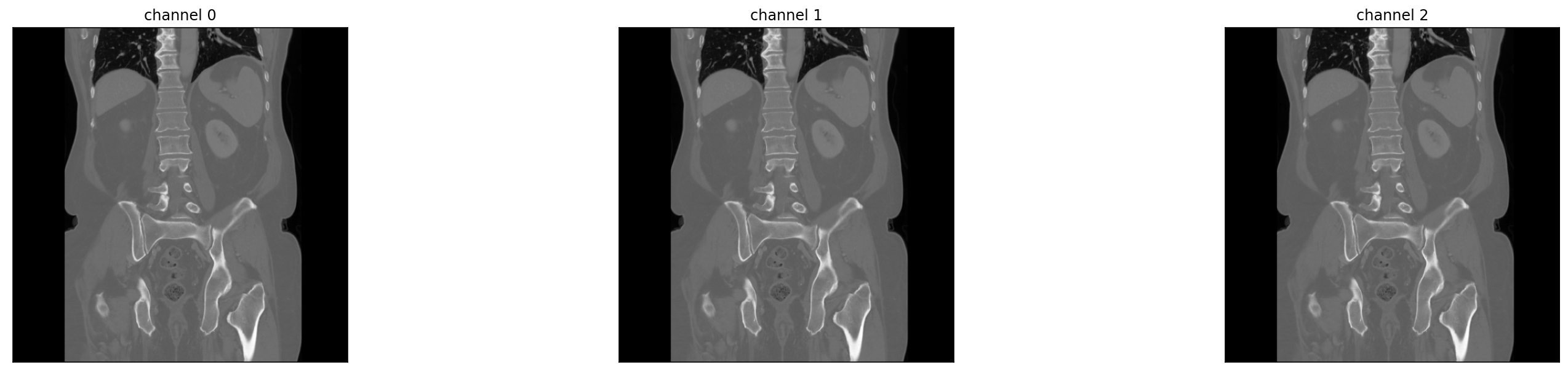
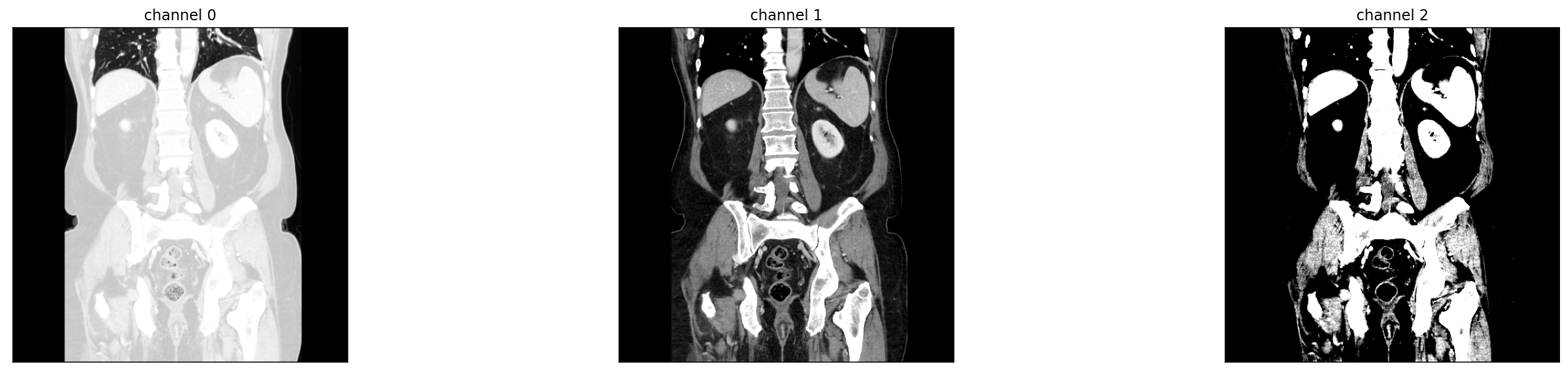
ds = radtorch.data.ImageDataset(folder='data/4CLASS/', out_channels=3, WW=[1500, 350, 80], WL=[-600, 50, 40])
ds.view_image(id=9)
VolumeDataset (Dataset)
Dataset object for DICOM Volume. Creates dataset(s) and dataloader(s) ready for training using radtorch or pytorch directly.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
folder |
str |
Parent folder containing images. |
required |
name |
str |
Name to be give to the dataset. If none provided, the current date and time will be used to created a generic dataset name. (default=None) |
None |
label_table |
str|dataframe |
The table containing data labels for your images. Expected table should contain at least 2 columns: image path column and a label column. Table can be string path to CSV or a pandas dataframe object.(default=None) |
None |
use_file |
bool |
True to use pre-generated/resampled volume files. To use Volume files:
|
False |
extension |
str |
Type/Extension of images. |
'dcm' |
out_channels |
int |
Number of output channels. |
1 |
WW |
int or list |
Window width for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
None |
WL |
int or list |
Window level for DICOM images. Single value if using 1 channel or list of 3 values for 3 channels. See https://radiopaedia.org/articles/windowing-ct. |
None |
path_col |
str |
Name of the column containing image path data in the label_table. |
'path' |
label_col |
str |
Name of the column containing label data in the label_table. |
'label' |
study_col |
str |
Name of the column containing study id in label_table. |
'study_id' |
transforms |
dict |
Dictionary of Albumentations transformations in the form of {'train': .. , 'valid': .. , 'test': .. }. See https://albumentations.ai/docs/getting_started/image_augmentation/. NOTE: If using already resampled/created volume files, transformation should be applied during volume creation not dataset i.e. Transforms specified here have no effect during training. |
None |
random_state |
int |
Random seed. |
100 |
sample |
float |
Sample or percent of the overall data to be used. |
1.0 |
split |
dict |
dictionary defining how data will be split for training/validation/testing. Follows the sturcture {'valid': float, 'test': float} or {'valid':'float'} in case no testing subset is needed. The percent of the training subset is infered automatically. |
False |
normalize |
bool |
True to normalize image data between 0 and 1. |
True |
batch_size |
int |
Dataloader batch size. |
16 |
shuffle |
bool |
True to shuffle images during training. |
True |
weighted_sampler |
bool |
True to use a weighted sampler for unbalanced datasets. See https://pytorch.org/docs/stable/data.html#torch.utils.data.WeightedRandomSampler. |
False |
num_workers |
int |
Dataloader CPU workers. |
0 |
Attributes:
| Name | Type | Description |
|---|---|---|
classes |
list |
List of generated classes/labels. |
class_to_idx |
dict |
Dictionary of generated classes/labels and corresponding class/label id. |
idx_train |
list |
List of index values of images/instances used for training subset. These refer to index of |
idx_valid |
list |
List of index values of images/instances used for validation subset. These refer to index of |
idx_test |
list |
List of index values of images/instances used for testing subset. These refer to index of |
table |
pandas dataframe |
Table of images , paths and their labels. |
table_train |
pandas dataframe |
Table of images used for training. Subset of |
table_valid |
pandas dataframe |
Table of images used for validation. Subset of |
table_test |
pandas dataframe |
Table of images used for testing. Subset of |
tables |
dict |
Dictionary of all generated tables in the form of: {'train': table, 'valid':table, 'test':table}. |
dataset_train |
pytorch dataset object |
Training pytorch Dataset |
dataset_valid |
pytorch dataset object |
Validation pytorch Dataset |
dataset_test |
pytorch dataset object |
Testing pytorch Dataset |
datasets |
dict |
Dictionary of all generated Datasets in the form of: {'train': Dataset, 'valid':Dataset, 'test':Dataset}. |
dataloader_train |
pytorch dataloader object |
Training pytorch DataLoader |
dataloader_valid |
pytorch dataloader object |
Validation pytorch DataLoader |
dataloader_test |
pytorch dataloader object |
Testing pytorch DataLoader |
dataloaders |
dict |
Dictionary of all generated Dataloaders in the form of: {'train': Dataloader, 'valid':Dataloader, 'test':Dataloader}. |
class_weights |
tensor |
Values of class weights, for imbalanced datasets, to be used to weight loss functions. See https://pytorch.org/docs/stable/generated/torch.nn.CrossEntropyLoss.html#torch.nn.CrossEntropyLoss. |
sampler_weights |
tensor |
Values of vlass weights, for imbalanced datasets, to be used to sample from the dataset using Pytroch WeightedRandomSampler. Affects only training dataset not validation or testing. See https://pytorch.org/docs/stable/data.html#torch.utils.data.WeightedRandomSampler |
Examples:
import radtorch
import albumentations as A
# Specify image transformations
T = A.Compose([A.Resize(256,256)])
# Create dataset object
ds = radtorch.data.VolumeDataset(
folder='data/PROTOTYPE/DIRECTORY/',
split={'valid':0.3, 'test':0.3},
out_channels=1,
transforms={'train':T,'valid': T,'test': T},
)
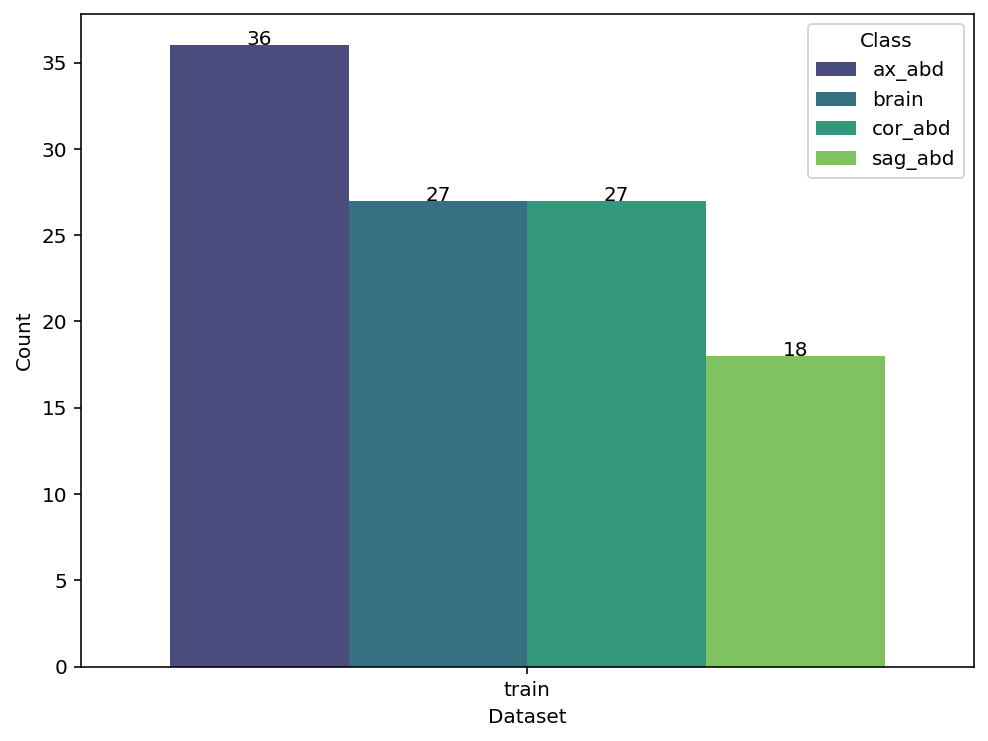
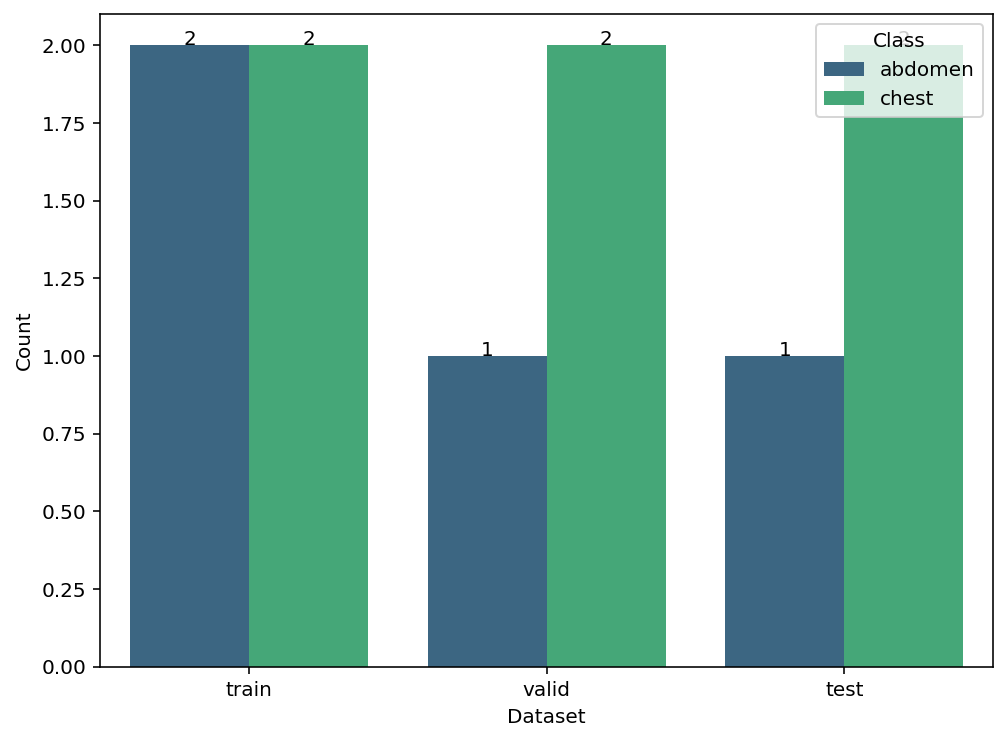
ds.data_stat()
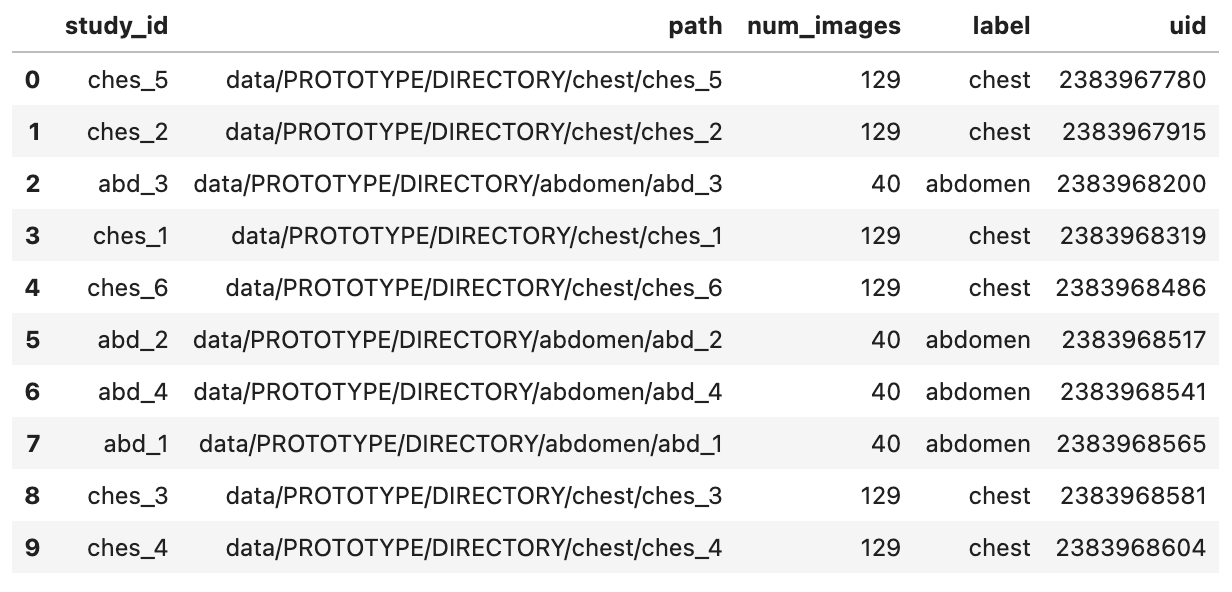
ds.table
Methods
data_stat(self, plot=True, figsize=(8, 6), cmap='viridis')
Displays distribution of classes across subsets as table or plot.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
plot |
bool |
True, display data as figure. False, display data as table. |
True |
figsize |
tuple |
size of the displayed figure. |
(8, 6) |
cmap |
string |
Name of Matplotlib color map to be used. See Matplotlib colormaps |
'viridis' |
Returns:
| Type | Description |
|---|---|
pandas dataframe |
if plot=False |
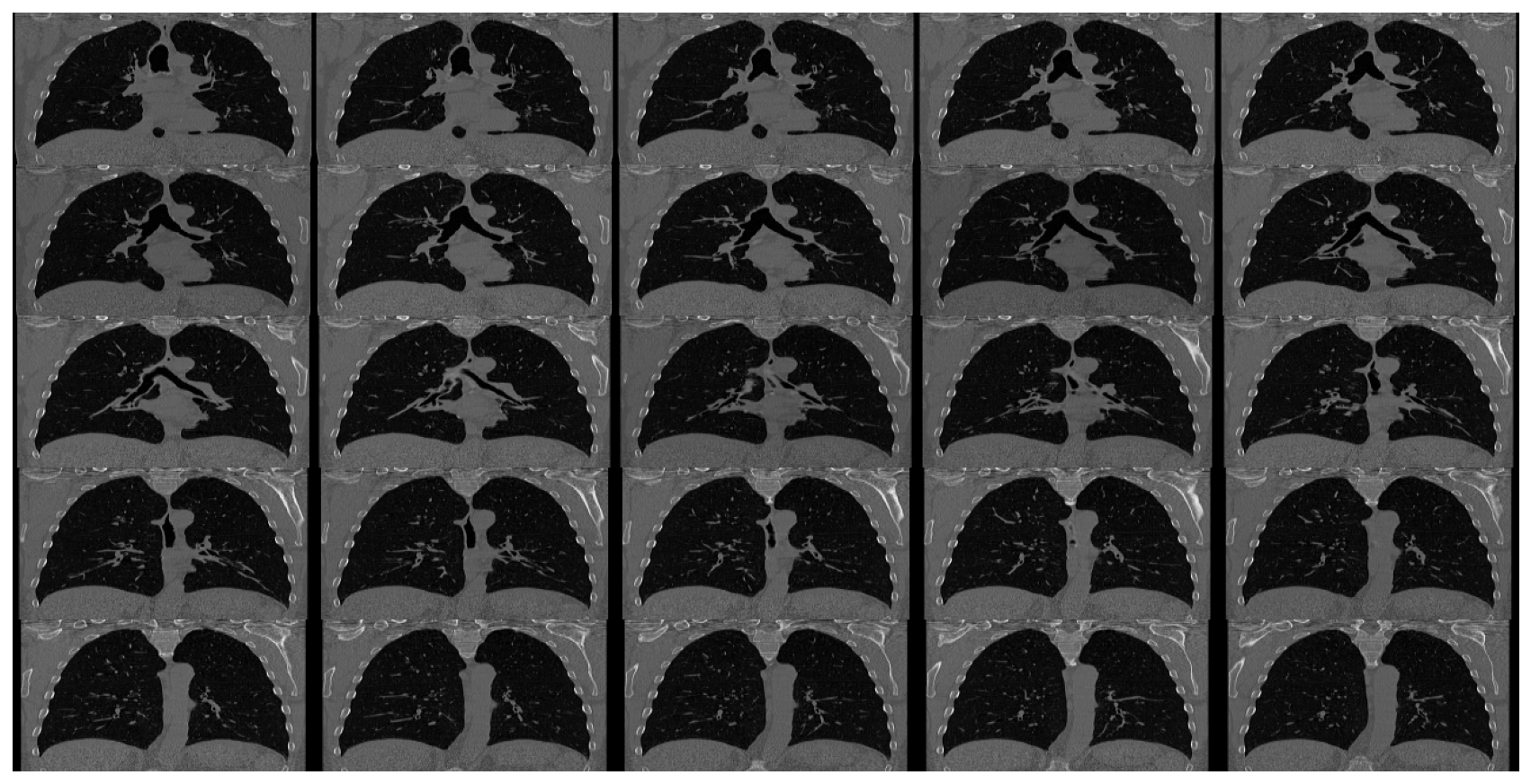
view_study(self, id, plane='axial', figsize=(15, 15), cols=5, rows=5, start=0, end=-1)
Show sample images from a study.
Warning
This works only with single channel images. Multiple channels are not supported yet here.
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
id |
int |
Target study id in |
required |
plane |
str |
Anatomical plane to display the images in. Options: 'axial', 'coronal' or 'sagittal'. |
'axial' |
figsize |
tuple |
Size of the displayed figure. |
(15, 15) |
cols |
int |
Number of columns. |
5 |
rows |
int |
Number of rows. |
5 |
start |
int |
id of the first image to display. |
0 |
end |
int |
id of the last image to display. |
-1 |
Returns:
| Type | Description |
|---|---|
figure |
figure containing images from study. |
Examples:
import radtorch
import albumentations as A
# Specify image transformations
T = A.Compose([A.Resize(256,256)])
# Create dataset object
ds = radtorch.data.VolumeDataset(
folder='data/PROTOTYPE/DIRECTORY/',
split={'valid':0.3, 'test':0.3},
out_channels=1,
transforms={'train':T,'valid': T,'test': T},
)
ds.view_study(id=0, plane='axial')
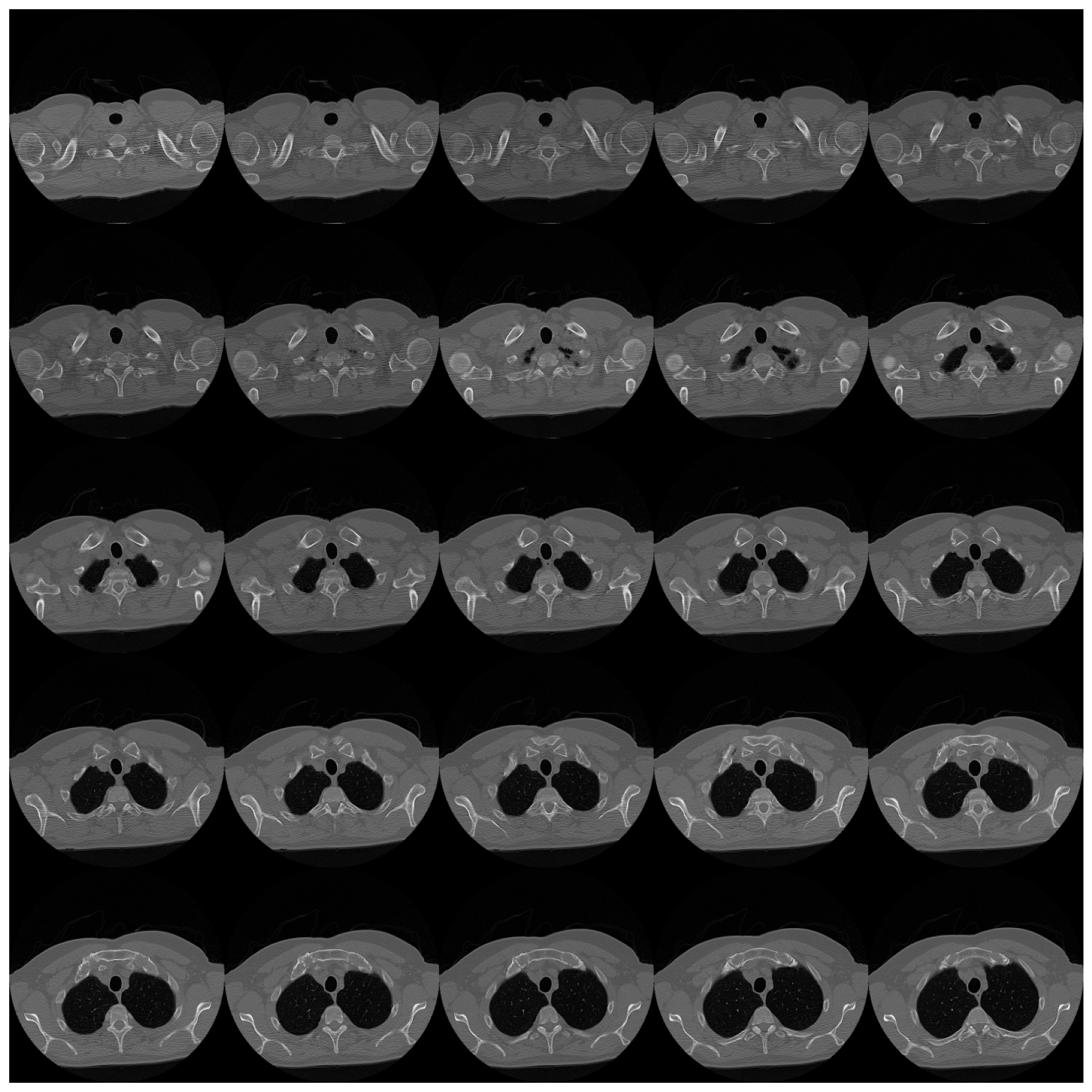
ds.view_study(id=0, plane='coronal', start=150)